PacBio Newsletter_Aug 2021
2021-08-24 09:43:30 Source:Gene Company Limited
Pacbio Update:
2021 APAC RARE & INHERITED DISEASE SMRT GRANT PROGRAM

WIN FREE SEQUENCING FROM A PACBIO SERVICE PROVIDER IN YOUR AREA
Sequence all life with confidence. With PacBio HiFi reads, you no longer need to compromise between long read lengths and high accuracy sequencing
Sequence all life with confidence. With PacBio HiFi reads, you no longer need to compromise between long read lengths and high accuracy sequencing
Submit Your Proposal For A Chance To Win Our SMRT Grant by submitting 300 words or less (or in 300 characters or less if submitting in Chinese) to shows how HiFi reads will accelerate your science.
Deadline: Thursday, 16 September 2021
Prize
- SMRTbell library construction (up to FOUR).
- PacBio sequencing (up to EIGHT 8M SMRT Cells).
- Preliminary bioinformatic analysis.
One winner will be selected from all qualified applications by a panel of PacBio scientists.
Learn more or apply at:
Co-sponsor:

MISS OUT the previous PacBio conference?
No Worry. Here is the chance to watch again on demand.
SMRT Leiden 2021 – Young Investigator Virtual Conference
About the event
The SMRT Leiden 2021 – Young Investigator Virtual Conference includes two back-to-back events: the 6th annual SMRT Scientific Symposium and the SMRT Informatics Developers Meeting.
This year SMRT Leiden will put the spotlight on young investigators who will share their latest scientific discoveries and novel analytical achievements using PacBio long-read sequencing.
On-demand sessions include:
- Presentations from PacBio users focused on key applications
- Live Q&A sessions and panel discussions to interact with speakers
- On-demand content for those new to PacBio
- Opportunity to connect with your peers in the Community
Please CLICK HERE to watch. If you have not registered yet, please register according to the website’s instruction prior to access the conference.
Document Update
July 29, 2021
Updated document to recommend Polymerase 2.2 + Primer v5 + Adaptive Loading Target = 0.85 + 2-hour Pre-Extension Time for HiFi De Novo/VD; Microbial Multiplexing; Low DNA Input; Ultra-Low DNA Input; and Shotgun Metagenomics applications. If using Binding Kit 2.0 for these applications, use a 2-hr Pre-Extension Time and do not enable Adaptive Loading.
July 29, 2021
Updated document to be consistent with latest sample setup and run
design recommendations described in the SQ II/IIe Loading & Pre-extension time QRC document.
June 30, 2021
Added the sentence “The three hifi_reads files are also available for download using Data > File Downloads in the same analysis” on Page 2.
July 21, 2021
New document
Video Update
Resolving Viral Evolution and Quasispecies Diversity with HiFi Sequencing
Ashby, Meredith; and Boritz, Eli
PacBio, National Institute of Allergy and Infectious Diseases
In this talk, speakers provide an understanding HiFi sequencing methods for resolving viral diversity in complex systems, examples of how HiFi sequencing can phase entire viral genes or genomes, revealing quasispecies diversity within patients, and how combining fully-phased minor variant data with other data types provides insights into viral evolution, immune escape, and drug resistance.
Optimizing for Information: What Richer Data and Better Assemblies Reveal About Metagenome Structure and Function
Ashby, Meredith; Bickhart, Derek, and Portik, Dan
PacBio, US Department of Agriculture
In this talk, speakers provide an overview of PacBio-recommended tools for metagenome sequencing analysis, where to download example test data, the typical performance for HiFi metagenome sequencing of fecal samples, the impact of read accuracy on metagenome assembly of long-read data, and finally, how deep sequencing that combines HiFi reads and Hi-C data can enormously increase recovery of high-quality MAGs and connect plasmids and viruses to host strains.
Whole-genome sequencing and analysis of two azaleas, Rhododendron ripense and Rhododendron kiyosumense
DNA Research. 2021 Jul 20.
- In this study, the whole-genome sequences of R. ripense is assembled by PacBio SMRT sequencing technology and that of R. kiyosumense is assembled by MGI single-tube long fragment reads (stLFRs) technology to enhance the phylogenetic analysis of azalea cultivars.
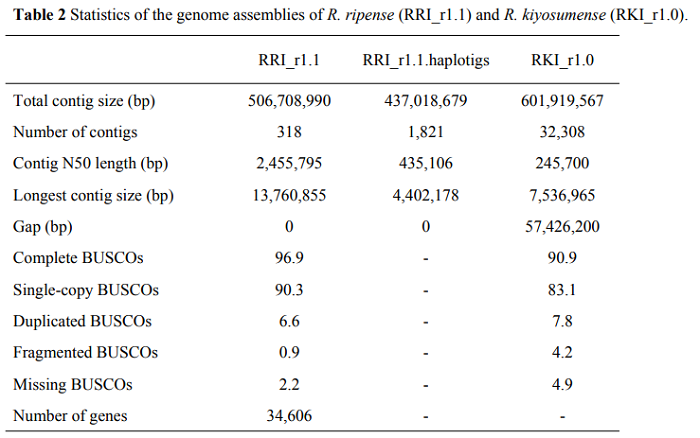
- From the table below , the Pacbio-sequenced R. ripense contigs (the assembled sequence contiguity represented by N50) was approximately 10 times longer than that of MGI-sequenced R. kiyosumense. The R. kiyosumense assembly is so fragmented with the number of contigs up to 100 times more than R. ripense and is not able to discern how many genes are in the genome. PacBio SMRT sequencing technology can get the two haplotypes separated but MGI stLFR technology can not.
- The difference in contig lengths between the two assemblies might be caused by the difference in the PacBio SMRT sequencing technology and the MGI stLFR technology. MGI stLFR technology actually involves the generation of short reads which are assembled together to form long single DNA molecules.
- PacBio SMRT sequencing technology now with HiFi reads can provide both long read lengths (up to 25 kb) and high accuracy (>99.9%) to quickly and affordably generate contiguous, complete, and correct de novo genome assemblies of even the most complex genomes.
Gene News





