PacBio Newsletter _Nov 2020
1970-01-19 21:53:52 Source:Gene Company Limited
Don’t miss out the chance to learn how HiFi sequencing can help you answer your toughest biological questions!
Please Register for PacBio Virtual Global Summit 2020 through the above “Register” button.
For the agenda, please click HERE for details.
2020 HiFi FOR ALL – Collaborations SMRT Grant Program

Virtual ASHG 2020 is over, and here are the links to access the videos of the PacBio workshop and CoLab. They are available on-demand, along with access to the various other assets.
•Workshop: https://programs.pacificbiosciences.com/l/1652/2020-09-29/427pvl
•CoLab: https://programs.pacificbiosciences.com/l/1652/2020-09-29/427tmn
•Posters: https://programs.pacificbiosciences.com/l/1652/2020-09-29/427tpv
•Literature: https://programs.pacificbiosciences.com/l/1652/2020-10-13/42b5v8

This Informational Guide provides detailed explanation of the sequencing accuracy. From this Guide, you can learn how HIFI reads with high accuracy generated from PacBio SMRT sequencing can achieve the coverage uniformity, mappability and phasing of the genome.
Link:

This new application note introduces the workflow of low and ultra-low DNA input for Whole genome sequencing with some case studies. With those workflow, samples with input DNA amounts as low as 5 ng can be applied and high-quality genome assemblies and human variant detection studies can be put within reach for many sample types that would otherwise be challenging to analyze using other long-read sequencing technologies.



•This study demonstrated the assemblies of reference-grade, phased diploid genomes of the cultivated apple (Malus domestica cv. Gala) and its two major wild progenitors, M. sieversii and M. sylvestris.
•The HIFI reads generated from PacBio Sequel II System were included in the assembly of diploid genome comprising phased scaffolds, as well as a conventional haploid consensuses, resulting in the final assemblies of diploid genome size of 1.31–1.32 Gb and high contiguity with the scaffold N50 of 3.3–4.3 Mb.
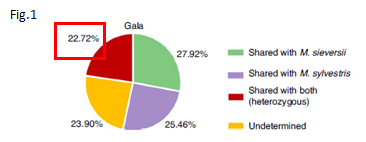
•The heterozygous Gala genome encoded around 23% of the sequence with hybrid ancestry, indicating that a considerable portion of the Gala genome has preserved genetic information from both progenitors. (Fig.1)
•Pan-genomes for the three Malus species were constructed separately by de novo assembly of the resequencing data, uncovering 1,736, 3,438 and 2,104 new genes were identified for M. sylvestris, M. sieversii and M. domestica. Hundreds of new genes being are selected from one of the progenitors and largely fixed in cultivated apples, revealing that introgression of new genes/alleles is a hallmark of apple domestication through hybridization.

AFA-enabled Epigenomic Applications
Optimizing ChIP Samples for Reproducibility (Tissues and Cells)
During this interactive seminar, we will highlight the key features and advantages of Covaris Adaptive Focused Acoustics® (AFA®) technology for Epigenetic workflows for cells, tissues, low input, Formalin-Fixed Paraffin- Embedded (FFPE) tissue, and native Chromatin Immunoprecipitation (ChIP).
•Simplify and standardize the complex ChIP sample preparation workflow
•Maintain epitope integrity during chromatin shearing
•Reduce tissue culturing cost and time by using fewer cells per experiment
•AFA-mediated IP reduces IP to 30 minutes from overnight
Date: 19 Nov 2020
Time: 9am (HK time)
Speaker

Hamid Khoja, Ph.D.,
Principal Scientist, Covaris, Inc.
Email: [email protected]
Please click HERE to register








