PacBio Newsletter (Issue Oct)
2020-10-16 15:11:00 Source:Gene Company Limited

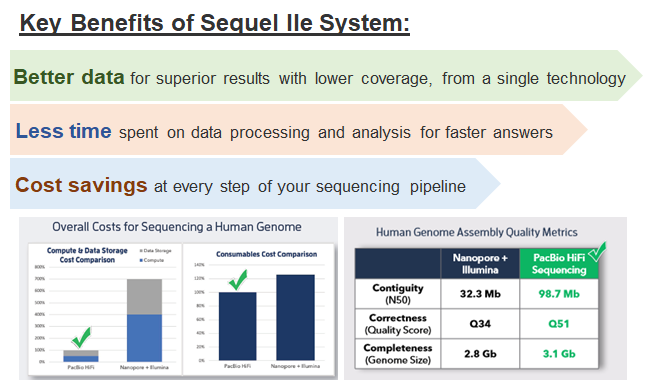
§Direct access to the only highly accurate long reads: PacBio HiFi reads
§Deeper biological insights, less data processing, and faster results thanks to the
unmatched clarity of HiFi reads
§Reliable and affordable high-throughput sequencing for a broad range of applications

v1.0 assembly of a complete human genome by The Telomere-to-Telomere (T2T) consortium
•Completion of all human chromosomes (all T2T) using PacBio HiFi reads, with the exception of the 5 rDNA arrays
•Assembly includes more than 100 Mbp of novel sequence compared to GRCh38
•High accuracy: Exceed Q60 (less than 1 error per million bases!)

For more details, please check HERE.
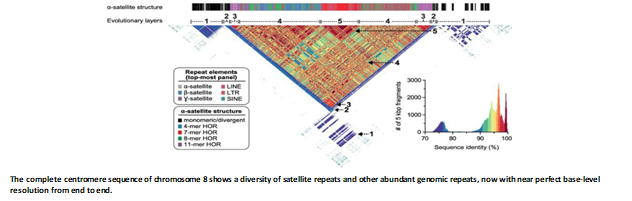
The First End-to-End Sequence of a Human Autosome, Chromosome 8
•The first completed autosome which is sequenced and assembled using PacBio HiFi reads
•A whole-chromosome assembly with an estimated base-pair accuracy exceeding 99.99%
•The sequence of five previously long-standing gaps is resolved, including a 2.08 Mbp centromeric α-satellite array, a 644 kbp defensin copy number polymorphism important for disease risk, and an 863 kbp variable number tandem repeat at chromosome 8q21.2 that can function as a neocentromere
For more details, please check HERE.


•Paper published in Nature Communication involves the using of HIFI reads for haplotype assemblies
•The benchmark covers 94%of the MHC locus and 22368 variants smaller than 50 bp. 49% more variants than a mapping-based benchmark using short read.
•This benchmark reliably identifies errors in mapping-based callsets, and enables performance assessment in regions with much denser, complex variation than regions covered by previous benchmarks
For more details, please check HERE.
New!
2020 Global Microbial Genomics SMRT Grant

SMRT Sequencing provides the highly accurate long reads, or HiFi reads, necessary to capture any microbe in high resolution.
Tell us in 250 words or less how SMRT Sequencing will drive new discoveries in your microbial research by November 6, 2020(Fri). You will stand a chance to win construction of up to 6 SMRTbell libraries, sequencing with up to 6 SMRT Cells 8M on the Sequel II System, and bioinformatics support.
Click HERE to apply now!

Protocol/document Updates
Protocol/document releases from Sep 2020.
Protocols | Released date | Changes |
Overview – Sequel Systems Application Options and Sequencing Recommendations | Sep 16, 2020 | 1.Fixed broken URL links to SQ/SQII QRCs and Application Consumable Bundle Purchasing Guide 2. Updated and clarified Minimum Recommended Coverage for each application type 3. Updated maximum recommended multiplexing level for No-Amp application from 10-plex to 20-plex 4. Updated list of recommended Data Analysis |
Sep 4, 2020 | New Release Application Note | |
Sep 4, 2020 | New guide to help troubleshoot sequencing performance issues on the Sequel II System | |
Quick Reference Card – Equipment and Materials Needed for Sequencing
| Sep 4, 2020 | 1.Updated application use case recommendations for the Genomic DNA 165 kb Kit (FP-1002-0275) and 55 kb BAC Analysis Kit (FP-1003-0275) for Femto Pulse systems from Agilent Technologies. 2. Updated equipment recommendations for Low DNA Input and Ultra-Low DNA Input applications |
Procedure & Checklist – Preparing HiFi SMRTbell Libraries from Ultra-Low DNA Input | Sep 4, 2020 | New Release Protocol |