PacBio Newsletter (Issue Sep)
2020-09-09 11:19:46 Source:Gene Company Limited

Pacbio Updates
PrecisionFDA Challenge
- PacBio HiFi Reads Outperform Both Short Reads and Noisy Long Reads
● Different NGS methods were evaluated for variant calling in human genomes, PacBio HiFi reads delivered the highest precision and recall in all categories: genome-wide, specifically in difficult-to-map regions, and in the major histocompatibility complex. (Fig. 1)
● Twenty-five of the 26 overall most accurate callsets used PacBio HiFi reads, including all of the top 12 (3 PacBio-only, 9 multi-technology) (circled in red in Fig 1).
● A submission from Google DeepVariant using PacBio HiFi reads had the fewest overall errors of any single-technology callset in the challenge. It had 5.8× fewer total errors than the popular combination of GATK with Illumina reads. (Fig. 2)
Fig. 1 Fig. 2
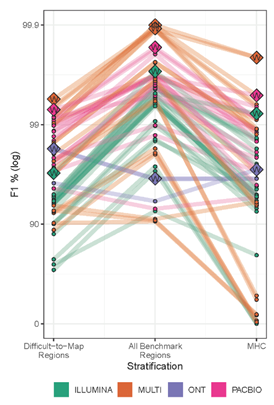
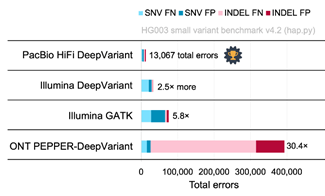
Total 64 entries: 17 using PacBio HiFi reads, 24 using Illumina reads, 3 using Oxford 
Nanopore reads, 20 using multiple technologies.
Article and publication Highlights


Articles in Clinical Omics (Vol 7, Issue No.4, Page 38-9) by Luke Hickey (Senior director of strategic marketing at PacBio)
◆ Short-read sequencing struggle to detect larger variants which cause rare diseases due to:
●Short reads containing non-unique sequence will map to many places in the genome, leading to assembly errors. For rare
diseases associated with repetitive regions e.g. ALS and Fragile X, mapping ambiguity and expansion length prevent
researchers from getting a clear view of the region of interest.
●Large variants cannot be fully spanned by short reads alone. It cannot be sequenced completely with breakpoints spanned and mapped for variant-calling algorithms to identify them correctly.
◆Long-read SMRT sequencing can produce highly accurate reads which are tens of kilobases long. Read with unique sequence are sufficient to capture even large variants in a single, uninterrupted readout.
◆SMRT sequencing has been used for identifying the structural variations causing rare disease like Carney complex, Epilepsy which cannot be found by short-read sequencer
◆SMRT sequencing is now being implemented in larger-scale efforts to solve rare diseases
e.g. European SOLVE-RD consortium and NIH’s Clinical Sequencing Exploratory Research program in US

Article and publication Highlights (cond’)

Tran et al., 2020, Mol Ther Methods Clin Dev. 18:639-651.
◆The gene therapy field has been galvanized by two technologies for genetic diseases: vectors based on adeno-associated viruses (AAVs), and CRISPR-Cas gene-editing tools. When combined into one platform, these safe and broadly tropic biotherapies can be engineered to target any region in the human genome to correct genetic flaws.
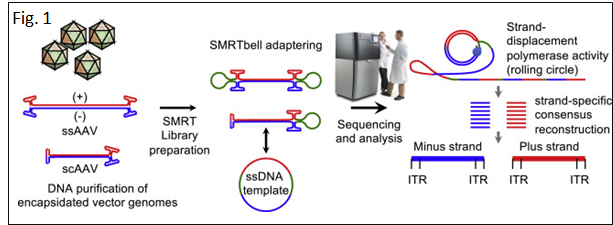
◆AAV-genome population sequencing (AAV-GPseq), a method based on PacBio SMRT sequencing (Fig. 1) previously revealed that self-complementary AAV vector designs with strong DNA secondary structures can cause a high degree of truncation events, impacting production and vector efficacy.
◆AAV vectors can package CRISPR components without compromising vector integrity. Using SMRT sequencing, it demonstrated that vector genome designs that carry dual sgRNA expression cassettes in tail-to-tail configurations lead to truncations.
◆Also, SMRT sequencing revealed the heterogeneity in inverted terminal repeat sequences in the form of regional deletions inherent to certain AAV vector plasmids.

Fast and easy micro-volume quantification of Nucleic acid in library preparation QC

Synergy Neo2 Hybrid Multi-Mode Reader
Take3 Micro-Volume Plate (used in Synergy Neo2)