PacBio Newsletter_Oct 2021
2021-10-14 00:00:00 Source:Gene Company Limited
Pacbio Update:
2021 Global Virtual Meeting
Date: Nov 9, 2021

About the event:
Highly accurate long-read sequencing, or HiFi sequencing, is being used around the world to inspire discoveries in human biomedical research, plant and animal sciences, and microbiology and infectious disease. This year, the Global Virtual User Meeting will showcase research using PacBio powered technology from whole genome sequencing for de novo assembly to comprehensive variant detection to RNA sequencing and more!
Register now for a 24-hour virtual journey to join fellow researchers and hear from experts to learn how HiFi sequencing is expanding knowledge of genomes and inspiring advancements in science. With three tracks running consecutively based on region, you have the flexibility to join us at your preferred time to watch. On the agenda page, simply filter by the region of your choice.
KEY SPEAKERS:

Christian Henry
Chief Executive Officer
PacBio

Dr. Jonas Korlach
Chief Scientific Officer
PacBio

Dr. Martin Mascher
Group Leader
Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben
Documents / Video updates
Sep 9, 2021
Updated step 13 on pages 23 and 24 to provide guidance on saving raw sequencing data.
Sep 15, 2021
Version 01. This document is for Customer IT or SMRT® Link Administrators, and describes the data files generated by Sequel II Systems and Sequel IIe Systems, and how to work with those files.
Long-Read Genome Sequencing for the Molecular Understanding of Rare Disease and Neurodevelopmental Disorders
Jonas Korlach, PacBio
Susan Hiatt, HudsonAlpha Institute for BioTechnology
Susan Hiatt discussed how she and her team used HiFi sequencing in their rare disease research to discover genomic variation missed by whole-exome or genome sequencing studies using short reads.
PacBio’s Chief Scientific Officer Jonas Korlach also provided a summary about how advancements in sequencing technology, coupled with improved analytical tools and databases, have made it possible to finally examine hard-to-reach regions of the human genome, with huge implications for rare disease research.
Revealing Mechanisms of Bacterial Virulence and Adaptation with PacBio SMRT Sequencing
Ashby, Meredith; Bickhart, Derek, and Portik, Dan
PacBio, Pairwise, and Brigham and Women's Hospital
In this talk, speakers will describe the importance of high accuracy and long read length for generating closed bacterial assemblies. Speakers will also share examples of how hard-to-assemble domains and plasmids impact important biological traits including, pathogen virulence and anti-microbial resistance. Finally, they will provide an overview of the advantages of highly accurate long-read sequencing for outbreak tracking.
Publication Highlights
NEW! Concatenation sequencing using HiFi
“PacBio sequencing output increased through uniform and directional fivefold concatenation”
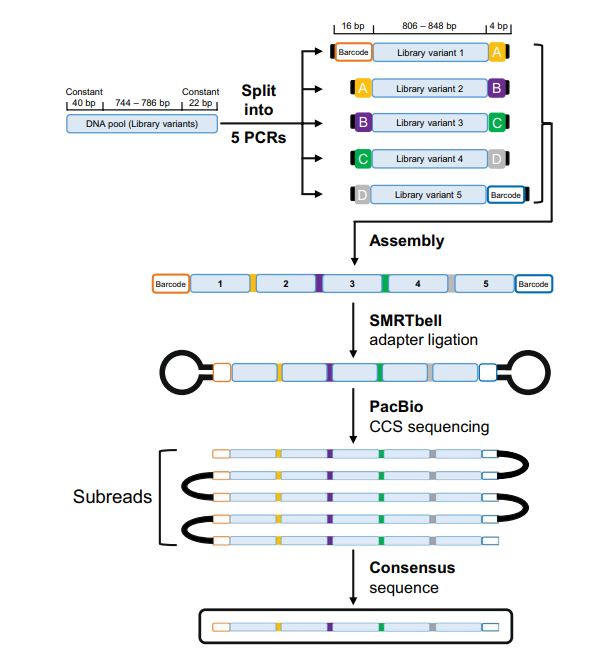
- In this paper, authors optimized a method to concatenate genes with ~870bp five times and then sequence the resulting DNA of ~5kb on PacBio Sequel system using CCS read.
- They also developed an all-in-one analysis pipeline, DeCatCounter, to take CCS sequencing read files (FASTA or FASTQ) as input for demultiplexing, deconcatenating, filtering by length and (optionally) translating DNA sequencing data to amino acid sequence.
- For an overview of the concatenation procedure, DNA pool to be sequenced was split into five PCR reactions with distinct primer sets, producing library variants 1–5.
- The PCR reactions also appended the variants with complementary BsaI restriction sites, which contained the four unique scar sequences (A–D) (yellow, purple, green, grey). The complementary overhangs are shown as black rectangles.
- Sample-specific barcodes were attached to the 5′-end of library variant 1 and the 3′-end of variant 5.
- The design enabled controlled and directional assembly of the five library variants upon Golden Gate assembly.
- Sequencing was carried out on a PacBio Sequel system, using one SMRT cell 1M and a collection time of ten hours. Circular consensus sequencing (CCS) was performed using the SMRT Link software v8.0.
- Sequencing data was filtered with at least 3 passes and a sequencing accuracy of at least 99% (Q20), which yielded a total of 124,715 sequences.
Gene News





